Holly Ellis, PhD
 Professor
Professor
Office 5S-32 (Laboratories 5S-30 & 5S-19)
Brody School of Medicine at East Carolina University
Greenville, NC 27834
phone: 252-744-1304
email: ellish20@ecu.edu
Education
- Wake Forest University School of Medicine, D., Biochemistry, 1998
- University of Central Florida, B.S., Microbiology, 1990
Professional Experience
- Brody School of Medicine, Professor, 2021
- NSF, Program Officer, 2011-2012
- Auburn University, Associate Professor, 2007-2016
- Auburn University, Professor, 2016
- Texas A&M University, NIH Post-Doctoral Fellow,1999-Aug. 2001
Research Interests
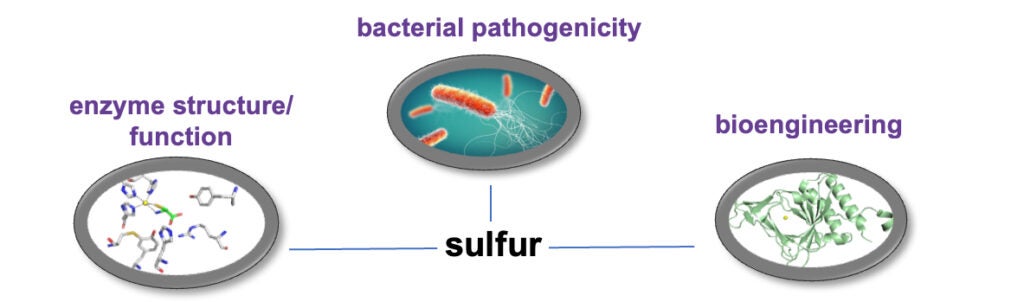
Sulfur is essential for all organisms as a component of amino acids and enzyme cofactors. Bacteria have developed extensive metabolic pathways to maintain cellular sulfur levels. Our lab has demonstrated the importance of sulfur availability for bacterial virulence and biofilm formation that can be exploited for drug development.
Our lab works on the structure/function of enzymes involved in sulfur metabolism. A combination of kinetic, spectroscopic, structural, and protein chemical methods are used by our lab to elucidate the catalytic mechanism and regulatory properties of these enzymes. Proteomic and metabolomic mass spectrometry are used to identify how metabolic pathways are affected during sulfur limitation.
Hydrogen Sulfide Metabolism in P. aeruginosa
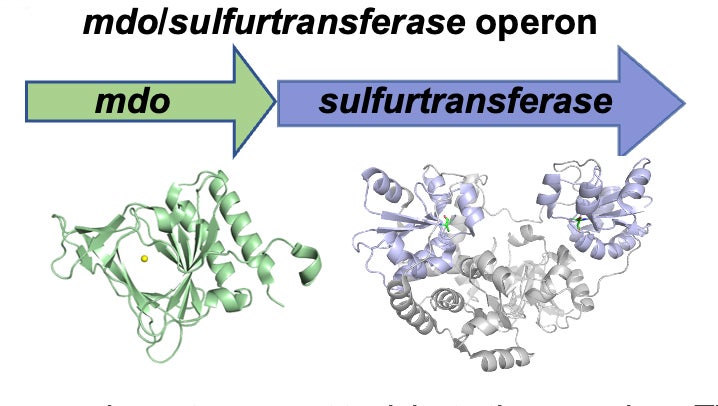 Pseudomonas aeruginosa is an opportunistic human pathogen that is especially problematic for individuals with cystic fibrosis and immunocompromised patients. P. aeruginosa evades host immune clearance through diverse metabolic processes. For example, hydrogen sulfide (H2S) protects P. aeruginosa from antibiotic-induced oxidative damage and host-produced reactive oxygen species. However, it is crucial that H2S concentrations are effectively maintained in P. aeruginosa to prevent toxicity to the organism. This combination of potential toxicity and utility of H2S presents an interesting metabolic challenge for the organism. Determining the mechanism involved in regulating H2S levels is critical to fully understand how P. aeruginosa can effectively utilize a potentially toxic virulence factor. The overall goal of these studies is to determine the mechanism and regulation of H2S metabolism in P. aeruginosa. Our group is evaluating the regulatory properties of the mdo/sulfurtransferase gene cluster in Pseudomonas aeruginosa and the mechanistic properties of the enzymes expressed from the operon.
Pseudomonas aeruginosa is an opportunistic human pathogen that is especially problematic for individuals with cystic fibrosis and immunocompromised patients. P. aeruginosa evades host immune clearance through diverse metabolic processes. For example, hydrogen sulfide (H2S) protects P. aeruginosa from antibiotic-induced oxidative damage and host-produced reactive oxygen species. However, it is crucial that H2S concentrations are effectively maintained in P. aeruginosa to prevent toxicity to the organism. This combination of potential toxicity and utility of H2S presents an interesting metabolic challenge for the organism. Determining the mechanism involved in regulating H2S levels is critical to fully understand how P. aeruginosa can effectively utilize a potentially toxic virulence factor. The overall goal of these studies is to determine the mechanism and regulation of H2S metabolism in P. aeruginosa. Our group is evaluating the regulatory properties of the mdo/sulfurtransferase gene cluster in Pseudomonas aeruginosa and the mechanistic properties of the enzymes expressed from the operon.
Sulfur acquisition in bacteria
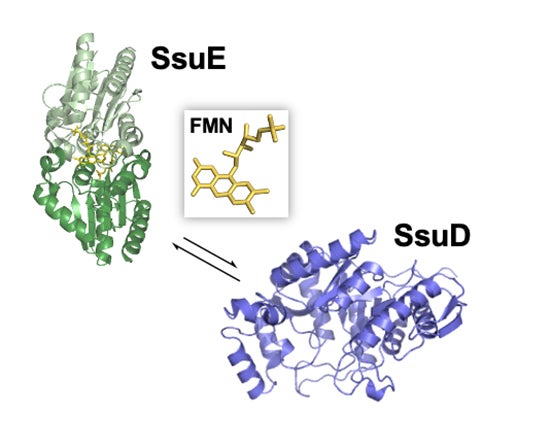 Sulfur metabolism in bacterial systems is tightly regulated to ensure that this critical element is available when standard sulfur sources are limiting. Bacteria typically rely on sulfate as their primary sulfur source but possess alternative pathways for sulfur acquisition when sulfate is limiting. Interestingly, several enzymes involved in sulfur acquisition are two-component FMN-alkanesulfonate dependent systems that rely on an FMN reductase and monooxygenase to catalyze the desulfonation of diverse organic sulfur compounds. In a diverse range of bacteria, these two-component systems expressed from different operons work together to catalyze the oxidation of environmental organic sulfur compounds. Combined structural and functional studies of these enzymes will provide insight into the mechanistic advantage of having a separate FMN reductase and monooxygenase enzyme to catalyze a single reaction, and how they work together to carry out their metabolic functions. Our recent investigations suggest that sulfur limiting conditions decrease bacterial virulence. Therefore, the alkanesulfonate monooxygenase enzyme would be a viable target for drug development. We are correlating the catalytic and structural features of these enzymes to understand the overall mechanism of desulfonation.
Sulfur metabolism in bacterial systems is tightly regulated to ensure that this critical element is available when standard sulfur sources are limiting. Bacteria typically rely on sulfate as their primary sulfur source but possess alternative pathways for sulfur acquisition when sulfate is limiting. Interestingly, several enzymes involved in sulfur acquisition are two-component FMN-alkanesulfonate dependent systems that rely on an FMN reductase and monooxygenase to catalyze the desulfonation of diverse organic sulfur compounds. In a diverse range of bacteria, these two-component systems expressed from different operons work together to catalyze the oxidation of environmental organic sulfur compounds. Combined structural and functional studies of these enzymes will provide insight into the mechanistic advantage of having a separate FMN reductase and monooxygenase enzyme to catalyze a single reaction, and how they work together to carry out their metabolic functions. Our recent investigations suggest that sulfur limiting conditions decrease bacterial virulence. Therefore, the alkanesulfonate monooxygenase enzyme would be a viable target for drug development. We are correlating the catalytic and structural features of these enzymes to understand the overall mechanism of desulfonation.
View complete bibliography: Holly Ellis